The IPCC at the University of Utah will focus on advancing the state-of-the-art in large-scale visualization through a combination of rendering and analysis techniques, using Intel Xeon and Xeon Phi architectures. I4DAV aims to advance large-scale and in situ visualization. With the majority of the top 10 (Top500 November 2013) supercomputers running SIMD CPU architectures, and the increasingly prohibitive cost of IO in high performance computing, CPU and manycore ray tracing technologies such as OSPRay, and streaming solutions such as ViSUS will be key to efficient large-scale visualization. Programming models such as IVL and ISPC enable performance on these architectures, and are increasingly useful for diverse visualization and HPC codes.
The IPCC at the University of Utah will focus on advancing the state-of-the-art in large-scale visualization through a combination of rendering and analysis techniques, using Intel Xeon and Xeon Phi architectures. I4DAV aims to advance large-scale and in situ visualization. With the majority of the top 10 (Top500 November 2013) supercomputers running SIMD CPU architectures, and the increasingly prohibitive cost of IO in high performance computing, CPU and manycore ray tracing technologies such as OSPRay, and streaming solutions such as ViSUS will be key to efficient large-scale visualization. Programming models such as IVL and ISPC enable performance on these architectures, and are increasingly useful for diverse visualization and HPC codes.
Research and Software
Intel OSPRay framework — Ray tracing for visualization
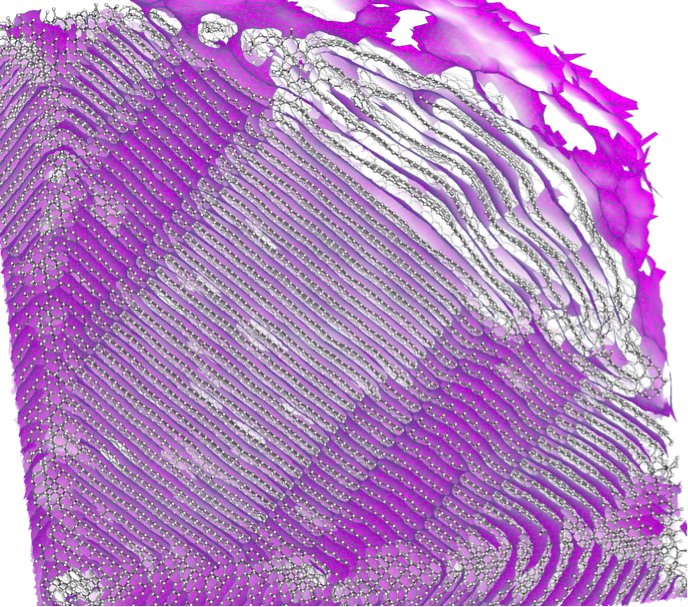
OSPRay is an open-source API developed and maintained by Intel, using the Embree ray tracing kernels and the Intel SPMD Program Compiler (ISPC. These ray tracing approaches allow small and large data to be rendered equally efficiently. I4DAV will bridge SCI Institute's expertise in particle data and volume rendering with this visualization framework from Intel.
http://ospray.github.io
ViSUS — Large-scale streaming visualization
Streaming methods such as ViSUS are necessary for visualization when data are so large that simplification is required for interactive exploration. ViSUS is a streaming level-of-detail (LOD) system for visualization of large image and volume data. It employs IDX, a lossless hierarchical data format for efficient streaming, analysis and visualization. ViSUS is designed for remote in situ visualization of extremely large (terascale--petascale) data, leveraging fast mechanisms for streaming data directly from simulation. Our research seeks to improve both ViSUS client and server technology with optimizations for CPU and manycore devices.
http://www.visus.org
IVL – Independent Vector Layer
Originally developed by Ingo Wald at Intel Labs, IVL is a research SPMD compiler similar to ISPC. In contrast to ISPC, IVL uses standalone wrappers which map SPMD code directly to SIMD independently from a precompiler. SCI has augmented the wrapper interface to support inclusion directly within C++ code, providing operator overloading and semantics for common SIMD control flow. IVL is currently maintained by SCI under the MIT license.
https://gforge.sci.utah.edu/gf/project/ivl/
Uintah — Heterogeneous Computational Framework
Uintah is a collection of simulation codes developed at the University of Utah, including continuum fluid dynamics (AMR) and solid mechanics (MPM particle) components, which can be efficiently executed together in parallel HPC environments using a heterogeneous scheduler. The driving problem for the use of Uintah under the PSAAP II grant is to simulate state-of-the-art electricity producing coal fired boilers, advancing research in clean coal and carbon capture techniques. I4DAV aims to optimize both computation and visualization of Uintah with Intel many-core architectures, and explore ways to optimize Uintah tasking and vectorization on Intel platforms.
http://www.uintah.utah.edu
Applications
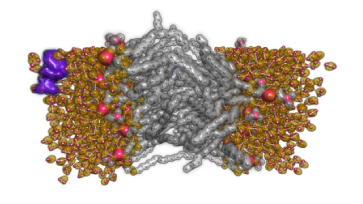
I4DAV targets a wide range of visualization and analysis problems through CEDMAV's partners in DOE national labs and industry. Applications include materials science and computational chemistry, combustion, cosmology, microscopy and tractography, and oil and gas.
Projects
- Large-scale particle visualization in OSPRay
- In situ visualization
- Analysis in materials science, particularly topological analysis leveraging multi-core and large-memory hardware.
- Optimizations to rendering and analysis in ViSUS, SCIRun and other frameworks.
- Research in improving programming models for efficient CPU visualization (IVL).
Related Publications
- Aaron Knoll, Ingo Wald, Paul Navratil, Anne Bowen, Khairi Reda, Michael E. Papka, Kelly Gaither. “RBF Volume Ray Casting on Multicore and Manycore CPUs”. Computer Graphics Forum (proc. Eurovis 2014), 3(33), 2014
- Aaron Knoll, Ingo Wald, Paul Navratil, Michael E Papka, and Kelly P Gaither. Ray Tracing and Volume Rendering Large Molecular Data on Multi-core and Many-core Architectures. Proc. 8th International Workshop on Ultrascale Visualization at SC13 (Ultravis), 2013.
- Ingo Wald, Aaron Knoll, Gregory P. Johnson, Will Usher, Valerio Pascucci and Michael E. Papka. “CPU Ray Tracing Large Particle Data with P-k-d Trees.” IEEE Visualization (SciVis) 2015 (conditionally accepted for publication).
- Pascal Grosset, Manasa Prasad, Cameron Christensen, Aaron Knoll and Charles D. Hansen. “TOD-Tree: Task-Overlapped Direct send Tree Image Compositing for Hybrid MPI Parallelism.” Eurographics Parallel Graphics and Visualization (EGPGV) 2015.
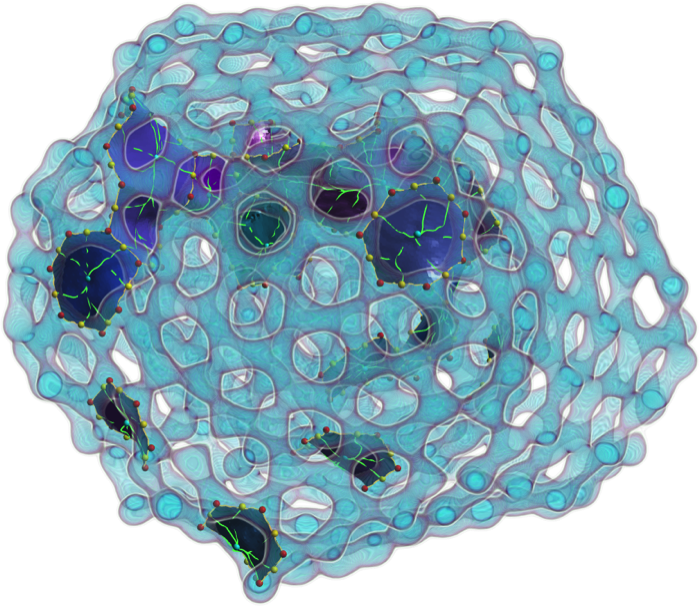
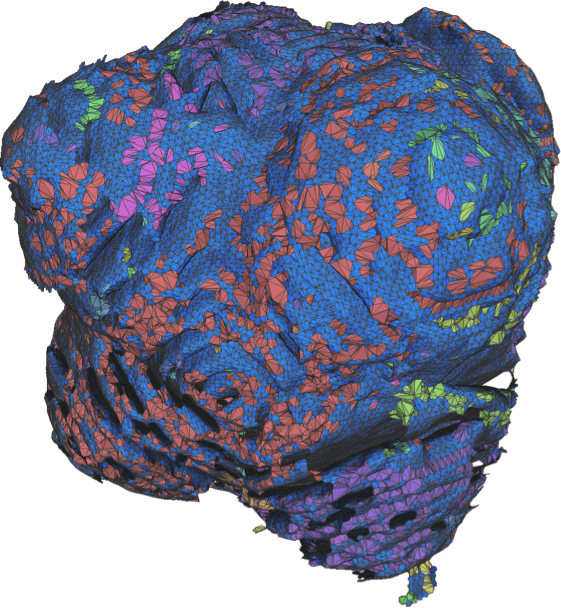
- Attila Gyulassy, Aaron Knoll, Kah Chun Lau, Bei Wang, Peer-Timo Bremer, Valerio Pasucci, Michael E. Papka and Larry Curtiss. “Morse-Smale Analysis of Ion Diffusion in Ab Initio Battery Materials Simulations.” Symposium on Topology in Visualization (TopoInVis) 2015.
- Attila Gyulassy, Aaron Knoll, Kah Chun Lau, Bei Wang, Peer-Timo Bremer, Valerio Pasucci, Michael E. Papka and Larry Curtiss. “Interstitial and Interlayer Ion Diffusion Geometry Extraction in Graphitic Nanosphere Battery Materials.” IEEE Visualization (SciVis) 2015 (conditionally accepted for publication).
 |
 |
|
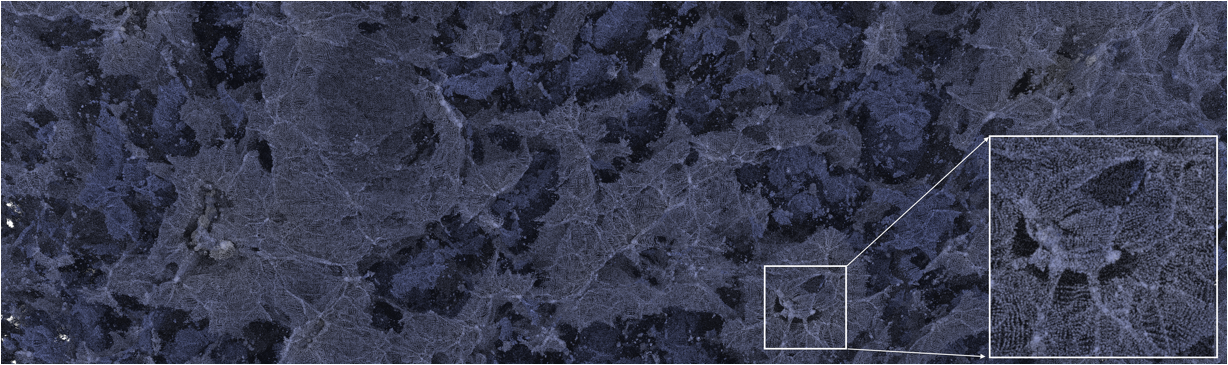
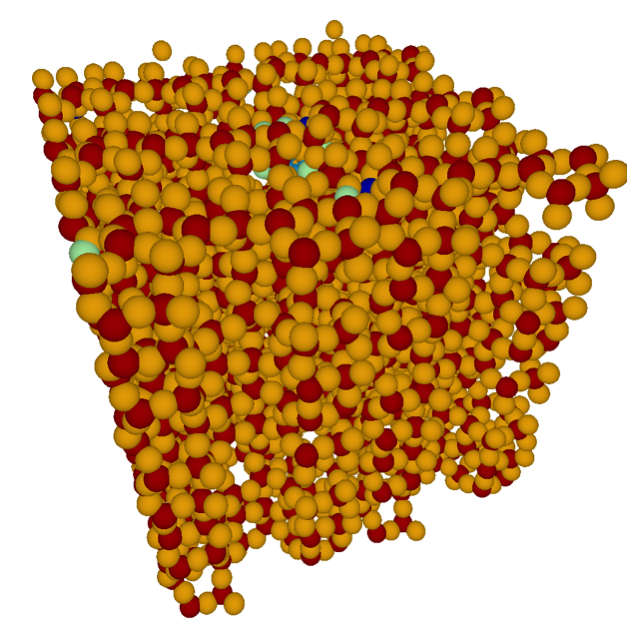
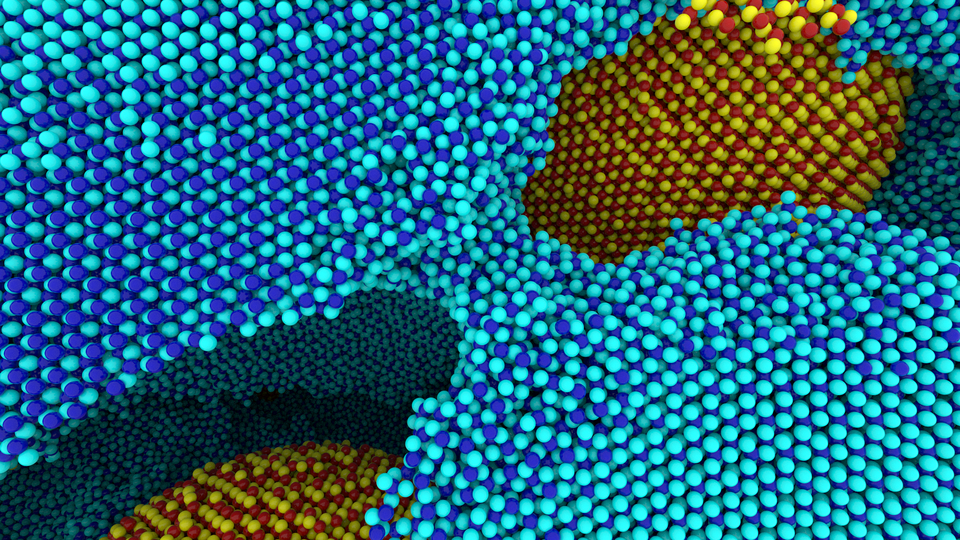
Figure 2: Path tracing a 100 million atom materials simulation conducted on Mira at Argonne National Laboratory; data courtesy Ken-ichi Nomura and Rajiv Kalia (University of Southern California). [3] |
|